Abstract
Drug resistance is a major obstacle in achieving complete and sustained therapeutic effect in cancer chemotherapy. Chemo-resistance may also lead to over-dosing and unwanted exposure to ineffective anti-tumor agents thereby increasing the risk of negative side-effects and the cost of drug development. Therefore, our goal is to utilize a large-scale pharmacogenomics database (Genomics of Drug Sensitivity in Cancer or GDSC, the largest public resource of drug-sensitivity data on over 250 drugs in more than 1000 human cancer cell lines of common and rare types of adult and childhood cancers of diverse origin) and develop a prediction method to identify novel secondary drug combination regimens that may effectively reverse drug resistance.
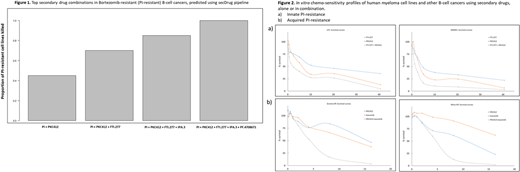
We utilized a greedy algorithm-based set-covering computational optimization method followed by a regularization technique to seek all secondary drugs that could kill maximum number of cell lines of the test disease (B-Cell cancers) resistant to the test drug (the Proteasome inhibitor/PI drug Bortezomib/Bz/Velcade) in a sequential manner ordered by the number of cell lines killed. The predicted top secondary drug combinations in PI-resistant B-cell cancers are shown in Figure 1.
To validate our prediction results, we treated human multiple myeloma cell lines (HMCLs) highly resistant to the proteasome inhibitors Bortezomib, Carfilzomob, Oprozomib and Ixazomib with the predicted best secondary drugs. Figure 2 depicts In vitro chemo-sensitivity profiles of PI-resistant HMCLs and other B-cell cancer cell lines showing percent survival compared to untreated control at increasing concentrations of secondary drugs, as single agents or in combination.
Furthermore, for each drug in the predicted drug combination, we identified differentially expressed (DE) genes by comparing the expression profiles between extraordinary- sensitive and resistant cell lines. These significantly regulated DE genes were used to identify pathways associated with the successful drug combinations.
Finally, we developed an R software package secDrug based on this computational pipeline for predicting novel secondary therapies in chemotherapy-resistant cancers. secDrug takes a query of any cancer type and any test drug, and outputs a list of the top secondary drug combinations with confidence score and biological pathway visualization. Thus, secDrug has potential application in clinical decision-making for discovering resistance-reversing cancer chemotherapy regimens.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal